17 Jul Taking a closer look at X-Ray Crystallography & X-Ray Diffraction at MU and MCW
Proteins come in numerous sizes and shapes. A protein’s structure is important in understanding its function. In order to determine the structure, researchers can use a technique called X-ray crystallography. Fundamentally, X-ray crystallography is a way to take pictures of proteins, thereby providing researchers with vital information regarding protein structure and ultimately function.
This technique utilizes an X-ray generator-area detector system of which there are only two in the Milwaukee area, at MCW and MU. MCW acquired a state-of-the-art diffraction facility a few years ago. However, after realizing that a single facility was insufficient to service all of the structural biologists in the Milwaukee area, in 2009 researchers applied to the National Science Foundation for the acquisition of a newer generation system. The grant application included collaborative proposals from investigators at MU, MCW, and UWM.
The Translational Technologies and Resources (TTR) of the CTSI was fortunate to have the opportunity to speak with both Martin St. Maurice, PhD, assistant professor of biological sciences and director of the X-ray crystallography facility at MU, and Jung-Ja Kim, PhD, professor of biochemistry and director of the X-ray crystallography facility at MCW, to get a better understanding of this unique technique and how it can be utilized for translational research studies.
What is high-intensity X-ray crystallography?
X-ray crystallography is the most common and powerful technique available for solving atomic structure of macromolecules. The method has been used to determine the structures of not only proteins but also biological macromolecules such as nucleic acids, carbohydrates, and lipids.
In this process, researchers purify a protein of interest, grow a protein crystal, and transmit a beam of X- rays at the protein crystal allowing it to diffract into many directions. The diffractometer then rotates the protein crystal while transmitting high energy X-rays onto it.
From the resulting data, a three-dimensional picture of electron density within the crystal is produced. This information allows researchers to generate a model detailing the atomic structure of the protein. Drs. St. Maurice and Kim both acknowledge that growing a protein crystal that works well is often adifficult task. It is an art with a little bit of luck!
What are the differences between the X-ray facilities at MCW and MU?
There are only a few differences between the X-ray facilities at the institutions. MU’s instrument is a slightly newer generation version, has higher intensity, and faster readout for screening a lot of crystals. MCW’s instrument has a larger detector, which has benefits with certain types of crystalmorphologies. MCW’s facility also offers high throughput automation to support protein crystallization, a necessary precursor to diffraction experiments.
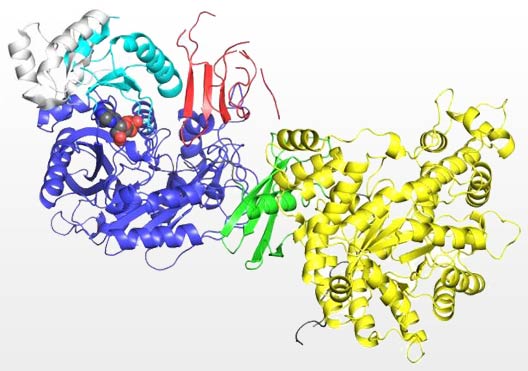
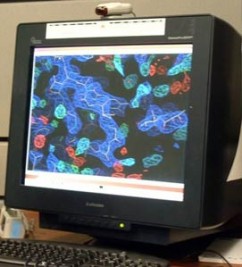
Pictured above is a crystal structure of a pyruvate carboxylase monomer color according to the individual protein domains. This structure, solved at Marquette University, was the first to reveal the molecular level interaction between the biotin carrier domain (red) and the biotin carboxylase domain (blue).
How can X-ray crystallography be used for clinical and translational research studies?
Projects involving X-ray crystallography are endless. Any research project that involves a macromolecule such as a protein, or cascade of proteins can benefit in some way from seeing what the structures look like. “X-ray crystallography allows us to go from a black box where we know very little about a particular protein to a point where we can get molecular level information and begin to understand how these proteins interact with other molecules”, said St. Maurice.[pullquote type=”2″ align=”right”]
“X-ray crystallography allows us to go from a black box where we know very little about a particular protein to a point where we can get molecular level information and begin to understand how these proteins interact with other molecules.”
– Martin St. Maurice, PhD
[/pullquote]
Dr. St. Maurice’s laboratory works at the exciting interface of chemistry and biology, using the tools of protein engineering, kinetic analyses and X-ray crystallography to determine the structure and function of various enzyme systems. Structural and kinetic analyses, used in tandem, provide a powerful means to probe underlying mechanisms of disease and unveil new targets for therapeutic applications.
The St. Maurice lab is seeking to determine the structures of multifunctional biotin-dependent enzymes with the goal of gaining insights into their basic function and applying these findings to the design of molecules that may be beneficial in thetreatment of type II diabetes, cancer and systemic infections.
Research labs at UW-Milwaukee are actively using the facilities to investigate the structure of enzymes implicated in antibiotic biosynthesis and enzymes that are targets for herbicides and therapeutics. Research into the structures of antibiotic producing proteins may one day allow these proteins to be modified to produce engineered antibiotics with altered or enhanced activities.
How can CTSI investigators access the facilities?
Various levels of training are available and collaborative arrangements can be made to scientists within the CTSI who wish to utilize the instruments.
Contact Information
MCW Facilities
Jung-Ja Kim, PhD
Email
MU Facilities
Martin St. Maurice, PhD
Email